By: Melissa Dellatorre
Hi, my name is Melissa and I am entering my second year as a graduate student in Dr. Donal Manahan’s lab at USC, where I use sea urchins and oysters to study early developmental physiology.
Two major goals of our lab are to increase the efficiency of the aquaculture business by studying influences on growth and development, and to determine how these species are likely to be affected by future global environmental changes such as temperature, food availability, and CO2.
My goals as a WIES Fellow for this summer are to further understand how food quantity and quality affects larval growth at the physiological, biochemical, and whole-organism levels. Although studies have been conducted for decades, there is still a large gap in knowing mechanistically how food is converted into energy, and how the energy budget of an organism is influenced by varying food concentration.

Left: Female Lytechinus pictus urchin spawning ~400,000 eggs into a small beaker. Middle: Aliquots of concentrated eggs for counting to determine total number. Right: Microscope used for counting and taking photographs used to measure size and growth.
This week my labmate Jason and I spawned white sea urchins, Lytechinus pictus, to create two million individuals. This ensures just enough biomass for a two-week experiment, assuming that all of the larvae develop properly. Once spawned, larval sea urchins develop quite rapidly through several phases. By four days old they reach a pluteus stage with developed digestive systems, and are ready to feed.
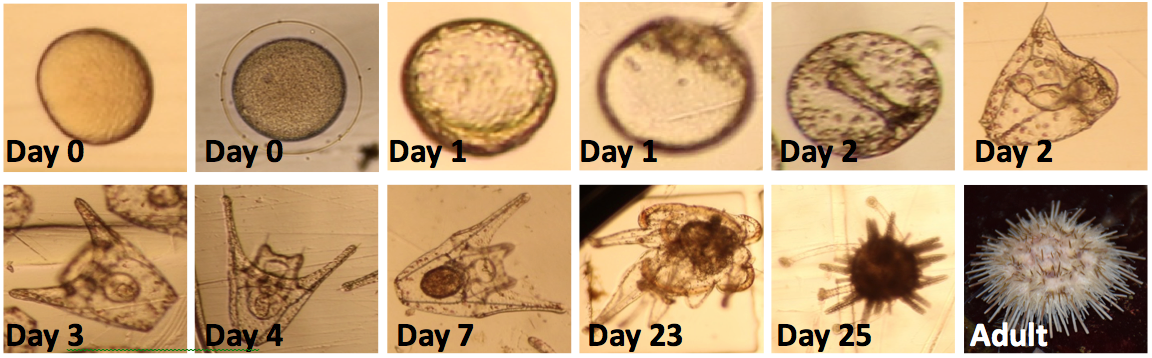
White sea urchin, Lytechinus pictus, development at 15°C. By day 1, organisms enter a blastula stage and begin to gastrulate. On day 2, they move from a gastrula to a prism. On day 3, pluteus stage is reached and their circular stomachs are evident. By day 25 some larvae have reached metamorphosis.
These two million individuals were allocated into six tanks, and are being fed a red alga, Rhodomonas lens, at high, medium, and low concentrations. From here, we take measurements to get the full picture of how these organisms are developing. This includes taking photographs to measure their growth, and doing biochemical analyses of protein, carbohydrate, and lipid contents of the algae and the larvae.
We also measure respiration and protein synthesis rates, as well as clearance rates which tells us how fast they are consuming their food. From this we can determine how many algae have been consumed by the larvae, giving us a measure of intake. This rate also tells us how often we need to top off the tanks with algae to keep food concentrations constant.

Experimental set up of our six 20 liter tanks (front row). Darker pigmentation can be seen in the tanks with the higher algal feeding concentrations.
My future experiments will compare this alga with other types of algae that white sea urchins like to eat, such as Dunaliella tertiolecta. The results can help shed light on food conversion efficiencies for white sea urchins, and determine the importance of nutrition during larval stages of development.
This research has implications for how larvae will fare under changing ocean conditions that affect food availability. It ultimately may also help to optimize the production of sustainable seafood for society.