By: Jordan Chancellor
Hello! My name is Jordan and I am now a second-year Ph.D. student at USC. My research focuses generally on population genomics in aquaculture species, which in my case is bivalves. My interest in bivalves, such as oysters and mussels, grew out of a combined passion for food security, nutrition, and sustainability. In my humble opinion, bivalves are the food of the future. They are packed with protein, serve as natural water filters, provide shelter to our delicate coastlines, and – most importantly – they’re delicious.
My original plan for this summer was to investigate sexual selection in oysters under the hypothesis that females are selecting mates based on chemical cues (termed “chemoattractants”) that their eggs sense in the water column, a novel occurrence unique to broadcast spawning organisms. I got scientific diving certified the past spring semester, and was looking forward to taking advantage of the facilities on Catalina, such as the Blue House, and having direct access to oysters housed on long lines in Cat Harbor. However, the shift to virtual research caused me to move that project to the back burner, and focus on something a little different.

Figure 2. Scientific diving in the Turks and Caicos Islands for the first time during my undergraduate study abroad program with The School for Field Studies. Hopefully more of this at Catalina in the future!
With research this summer being online, I started thinking more deeply about another novel observation in oysters that members of my lab have been working on for a while now. Nate Churches Ph.D., has calculated the generational per nucleotide mutation rate in Pacific oysters at 10e-5. While this number may seem arbitrary, keep in mind that in humans (and most other species for that matter) this value is approximately 10e-9. No mutation rate this high has ever been observed in eukaryotes until now! With this new knowledge in the back of my mind, I started to question what exactly was going on in the oyster genome that may be driving this observed mutation rate, and what consequences this may have on the genomic landscape as a whole.
To further investigate the oyster genome, I began employing the SLiM software, a forward-in-time evolutionary simulation framework that was developed by the Messer Lab at Cornell University (https://messerlab.org/slim/). I taught myself how to use the software in order to simulate chromosomal changes in the oyster genome over time, given the specific set of parameters I fed into it. These parameters include statistics like recombination rate, larval survival, reproductive success, number of generations, and our already empirically calculated mutation rate. Given these parameters (and a lot of coding), I have successfully obtained some preliminary data that gives us insight into the oyster genome.
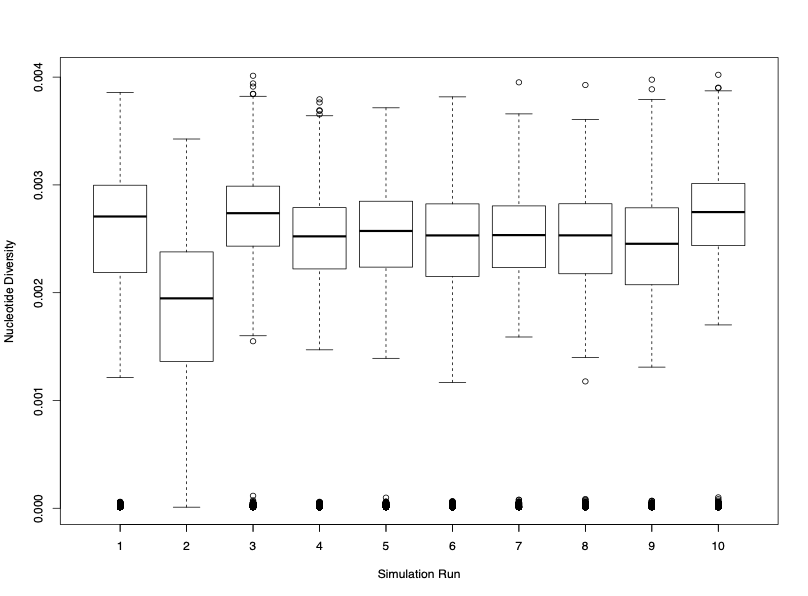
Figure 3. Boxplots of nucleotide diversity across the oyster chromosome from ten preliminary replicate SLiM simulation runs.
My first objective was to attempt to simulate the observed nucleotide diversity (π) value given our input parameters. The first set of simulations, however, were an order of magnitude smaller than empirically calculated values (0.003 versus 0.013; Zhang et al., 2012), informing me that we need to alter the simulations to better align with the oyster genome. Some of these alterations include: increasing the number of simulation replicates, increasing the number of generations the simulation is allowed to run for, and inputting the full range of calculated mutation rates in order to see if we can get closer to our expected π values. Along with making these changes to the simulation, I plan to conduct at least 1,000 replicate simulations in order to grasp the full scope of the genomic landscape of oysters and gain more statistical power. Using this type of information, we can hopefully inform future directions for hatchery and breeding practices for an efficient, safe, and sustainable aquaculture food product.